Use case #02: Case, light sensitivity
Open and reproducible analysis of light exposure and visual experience data (Advanced)
1 Preface
A single patient reports worse sleep after bright days. Consequently, personal light exposure and activity was captured with wearable devices for several weeks. Additionally, morning and evening questionnaires were used to capture sleep, performance, and well-being self evaluations. In this tutorial, we focus on the ActLumus device worn on the chest and the self evaluations.
The tutorial focuses on
import and merging of diary data with light data
calculation of metrics based on sleep-wake cycles instead of calendar days
relating exposure metrics to self assessments of consecutive sleep, performance, and wellbeing
assessing compliance to Brown et al. (2022)1 recommendations
2 How this page works
This document contains the script for the online course series as a Quarto script, which can be executed on a local installation of R. Please ensure that all libraries are installed prior to running the script.
If you want to test LightLogR without installing R or the package, try the script version running webR, for a autonymous but slightly reduced version. These versions are indicated as (live), whereas the current version is considered (static), as it is pre-rendered.
To run this script, we recommend cloning or downloading the GitHub repository (link to Zip-file) and running the respective script. You will need to install the required packages. A quick way is to run:
3 Recording
You can follow along with the recordings to get even more context about the workflow in LightLogR.
4 Setup
We start by loading the necessary packages.
5 Import
We require both light and log data to be loaded into R before we are able to merge them.
5.1 Light
Light data were captured with ActLumus devices. The exported data files sit in data/case_light_sensitivity/.
file <- "data/case_light_sensitivity/Log_5153_20251028143545799.txt"
tz <- "Europe/Berlin"
coordinates <- c(48.775555555556, 9.1827777777778247) #Stuttgart
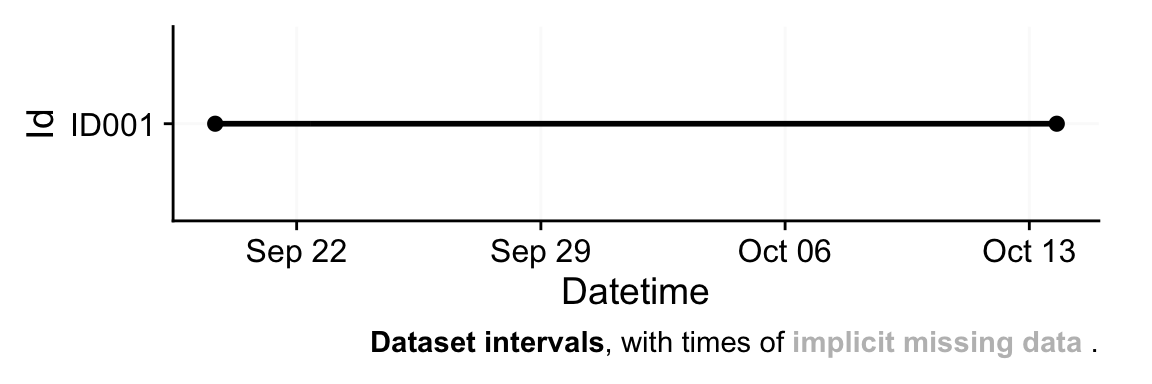
1data <- import$ActLumus(file, tz = tz, manual.id = "ID001")- 1
-
As there is only one participant, we set a manual
Id
Successfully read in 34'735 observations across 1 Ids from 1 ActLumus-file(s).
Timezone set is Europe/Berlin.
First Observation: 2025-09-19 15:57:16
Last Observation: 2025-10-13 18:53:31
Data from before 2001-01-01 were not imported. Adjust with `not.before` if needed.
Timespan: 24 days
Observation intervals:
Id interval.time n pct
1 ID001 60s (~1 minutes) 34733 100%
2 ID001 195s (~3.25 minutes) 1 0% This dataset spans about three weeks. We know that the diaries only start on 24 September 2025 - thus we remove prior data. We also test for irregular data or gaps in this trimmed dataset.
- 1
-
If we have a hard-set beginning or end time,
filter_Date()orfilter_Datetime()provide convenient cut-off functionality. - 2
-
As we are dealing with a single participant, we don’t require grouping by
Id.
[1] FALSE
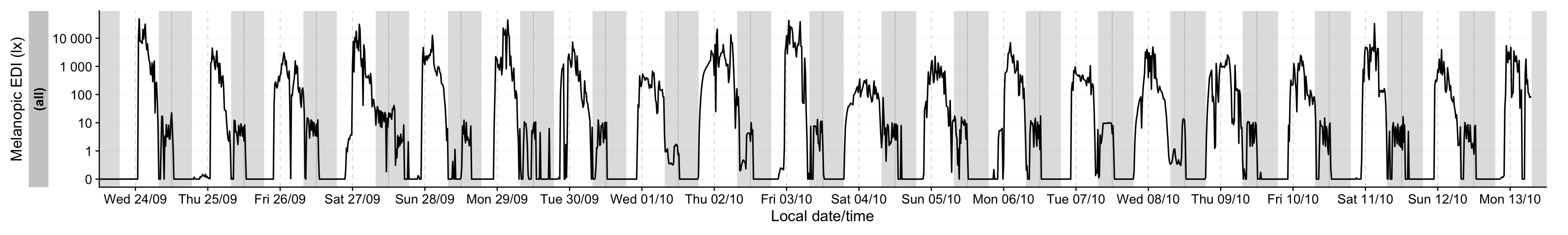
[1] FALSEWe can continue seeing as there are no issues with the regular time series. We start by plotting the full series
- 1
- Reduce the interval by averaging over 15 minute periods
- 2
-
Easily change the label format. Look at the help page of
?strptimefor the available syntax.
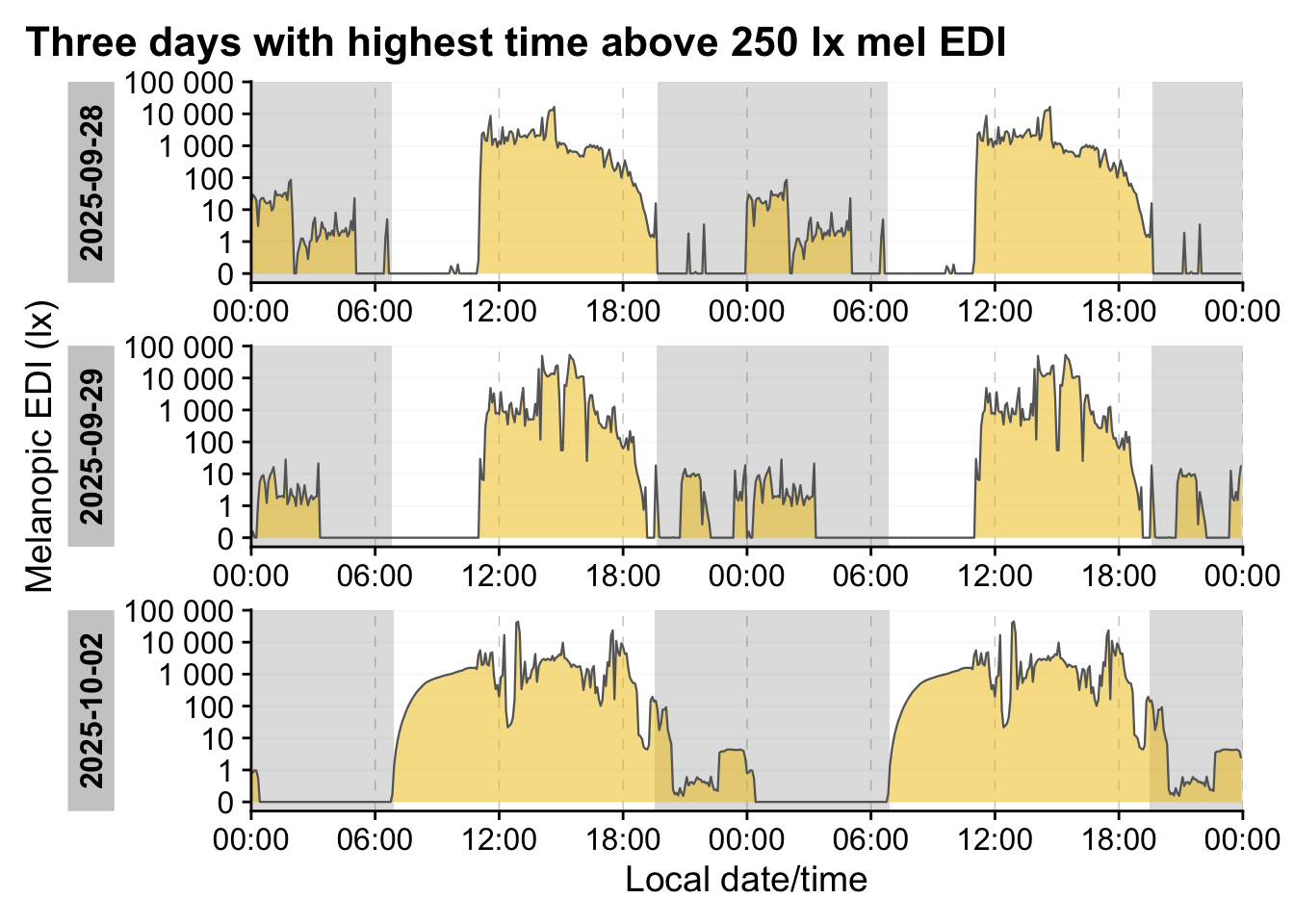
We can see the light intensity varies greatly. Let’s look at the extremes.
data|>
aggregate_Datetime("5 minutes") |>
1 add_Date_col(group.by = TRUE) |>
2 sample_groups(n=3,
order.by =
duration_above_threshold(MEDI, Datetime, threshold = 250)
) |>
ungroup(Date) |>
gg_doubleplot(type = "repeat") |>
gg_photoperiod(coordinates) +
labs(title = "Three days with highest time above 250 lx mel EDI")- 1
- Adding, and grouping by, the date gives us 20 groups, each covering one calender day
- 2
- Select the three groups with the highest duration above 250 lx mel EDI
Looking at the first half of 02 October 2025, we can already see that this likely a period of non-wear, or sleep - this, we have to come back to later.
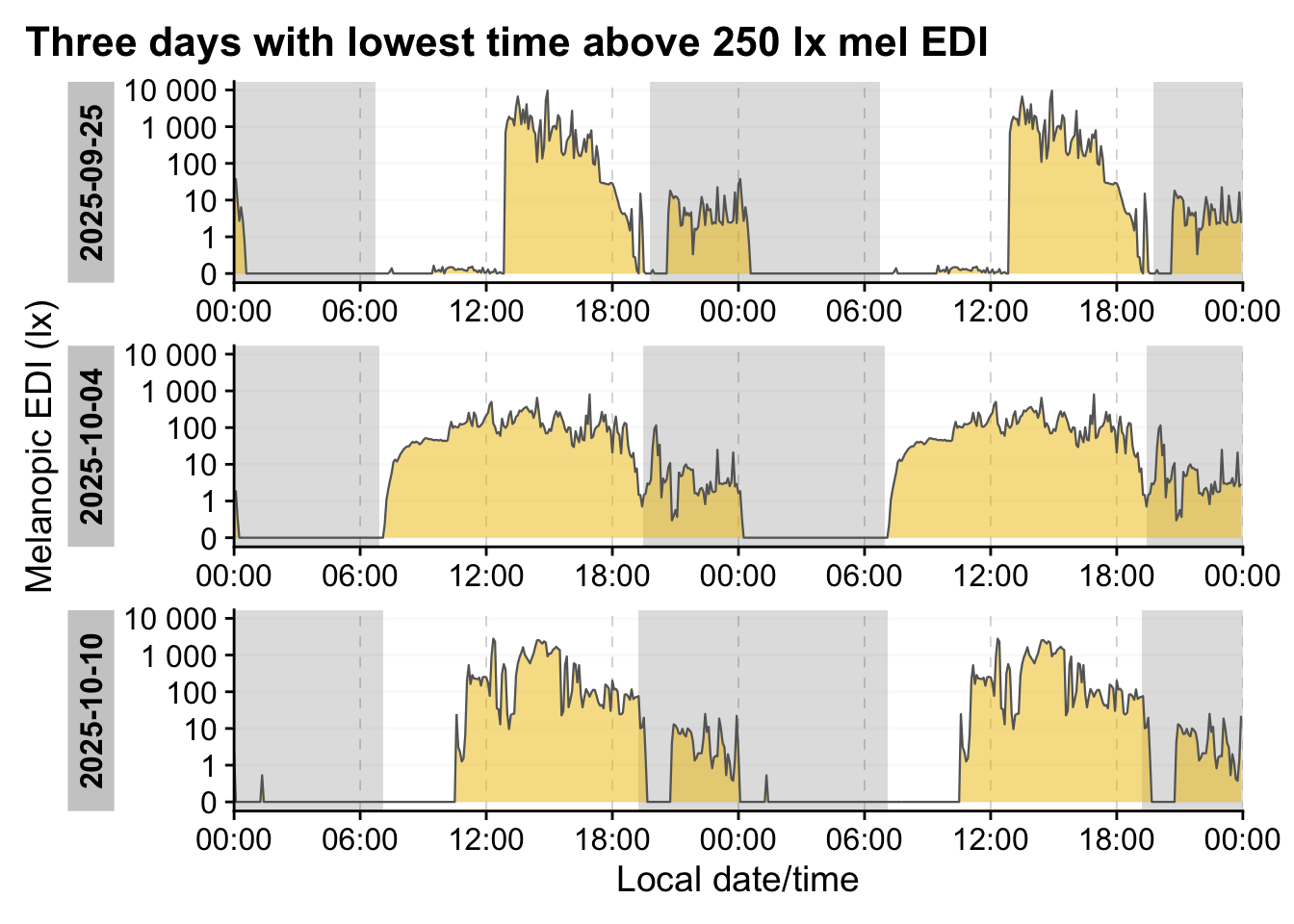
data|>
aggregate_Datetime("5 minutes") |>
add_Date_col(group.by = TRUE) |>
sample_groups(n=3,
1 sample = "bottom",
order.by =
duration_above_threshold(MEDI, Datetime, threshold = 250)
) |>
ungroup(Date) |>
gg_doubleplot(type = "repeat") |>
gg_photoperiod(coordinates) +
labs(title = "Three days with lowest time above 250 lx mel EDI")- 1
- Select the three groups with the lowest duration above 250 lx mel EDI
5.2 State data
We need information about sleep times. These can be found in the diaries.
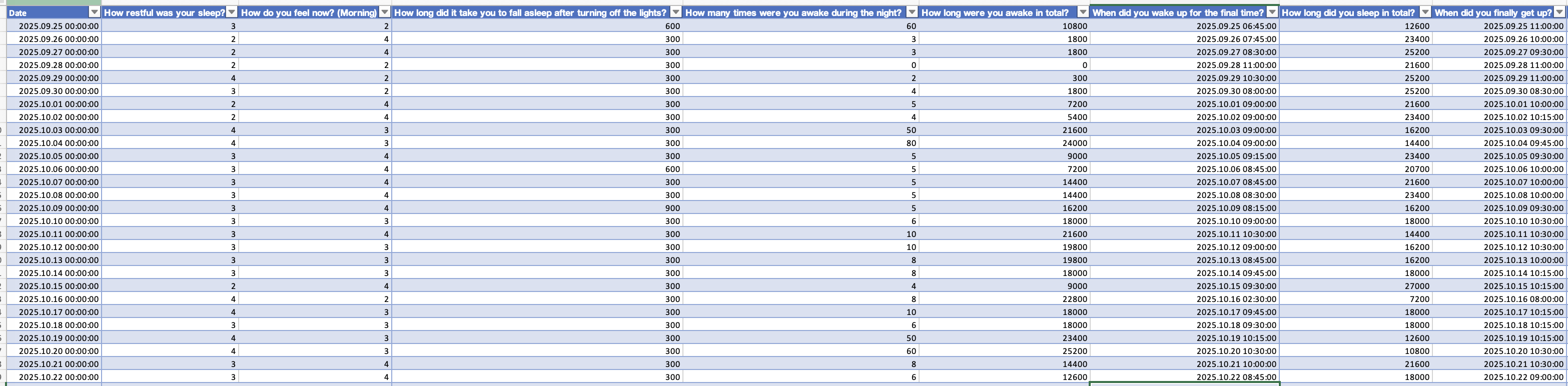
The morning diary is all about last nights sleep, how long the needed to fall asleep, wake-ups during the night, when they got up, how rested they feel and about sleep medication. See Figure 2 for an excerpt.
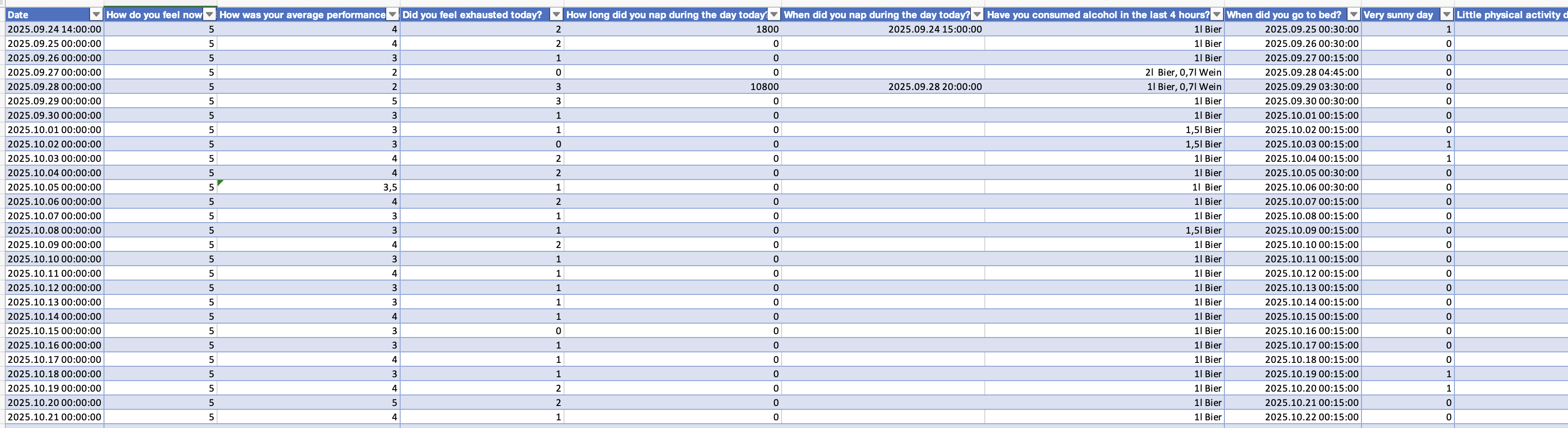
The evening diary is about the prior day, how well and they feel and about their performance during the day, alcohol consumation, when they got to bed, daytime naps and self-assessments about low activity and bright days. See Figure 3 for an excerpt.
In the next step we import these data and combine them to a large table. While none of these steps contain LightLogR functions, it is a crucial step to set the data up right for merging - particularly in that we assign the morning diaries to the prior day, and also add information about performance and fatigue on a given day to the prior day.
state_path1 <- "data/case_light_sensitivity/evening_protocol.xlsx"
state_path2 <- "data/case_light_sensitivity/morning_protocol.xlsx"
state_evening <- read_excel(state_path1)
state_morning <- read_excel(state_path2)
1state_evening$Date <- date(state_evening$Date)
state_morning$Date <- date(state_morning$Date - days(1))
2states <- left_join(state_evening, state_morning, by = "Date")
labels_states <-
names(states) |>
set_names(c("Date", "wellbeing.evening", "performance", "fatigue",
"daysleep.duration", "daysleep.start", "alcohol", "bedtime",
"sunny.day", "low.movement",
"sleepquality", "wellbeing.morning", "sleepdelay",
"sleepinterruptions", "sleepinterruptions.duration", "wakeup",
"sleepduration", "outofbed", "medication"))
names(states) <- names(labels_states)
states <-
3 states |> mutate(performance.next = lead(performance),
fatigue.next = lead(fatigue))- 1
- Store the date information of the evening diaries as is, but the morning diary will be set one day back.
- 2
- By joining these two diaries by their (adjusted) date, morning diary information is related to the prior day evening diary.
- 3
- We add the next days performance and fatigue level to all days.
The following table gives a short overview of the states. We will later get to whether a higher rating is better or worse for the individual items.
| Characteristic | N = 281 |
|---|---|
| How do you feel now? (Evening) | |
| 5 | 28 (100%) |
| How was your average performance today? | |
| 2 | 2 (7.1%) |
| 3 | 12 (43%) |
| 3.5 | 1 (3.6%) |
| 4 | 11 (39%) |
| 5 | 2 (7.1%) |
| Did you feel exhausted today? | |
| 0 | 3 (11%) |
| 1 | 15 (54%) |
| 2 | 8 (29%) |
| 3 | 2 (7.1%) |
| How long did you nap during the day today? | |
| 0 | 26 (93%) |
| 1800 | 1 (3.6%) |
| 10800 | 1 (3.6%) |
| Very sunny day | 5 (18%) |
| Little physical activity during the day | 5 (19%) |
| Unknown | 1 |
| How restful was your sleep? | |
| 2 | 6 (21%) |
| 3 | 15 (54%) |
| 4 | 7 (25%) |
| How do you feel now? (Morning) | |
| 2 | 5 (18%) |
| 3 | 10 (36%) |
| 4 | 13 (46%) |
| How long did it take you to fall asleep after turning off the lights? | |
| 300 | 25 (89%) |
| 600 | 2 (7.1%) |
| 900 | 1 (3.6%) |
| How many times were you awake during the night? | 6 (5, 10) |
| How long were you awake in total? | 14,400 (7,200, 19,800) |
| How long did you sleep in total? | 18,000 (16,200, 23,400) |
| Have you taken any sleep medication since yesterday evening? | |
| 0 | 28 (100%) |
| Performance next day | |
| 2 | 2 (7.4%) |
| 3 | 12 (44%) |
| 3.5 | 1 (3.7%) |
| 4 | 10 (37%) |
| 5 | 2 (7.4%) |
| Unknown | 1 |
| Fatigue next day | |
| 0 | 3 (11%) |
| 1 | 15 (56%) |
| 2 | 7 (26%) |
| 3 | 2 (7.4%) |
| Unknown | 1 |
| 1 n (%); Median (Q1, Q3) | |
6 Merging sleep data
Now that the diaries are prepared, we can extract sleep data from the states. The goal for the next code cell is to end with a statechange variable, that has a datetime for every timepoint when a new state occurs.
- 1
- Calculate sleep timing by adding bedtime and sleepdelay
- 2
- Reduce the dataset to only sleep and wake times
- 3
- We transform the data from wide to long format (i.e., consecutive wake-sleep statesd)
- 4
-
We add an initial
wakefor the first day
While we are at it, we can also create states according to the Brown et al. (2022) recommendations, which distinguish daytime (wake), evening (pre-sleep), and nighttime (sleep) hours. The LightLogR function sleep_int2Brown() makes this conversion a breeze. By default, the evening phase is set to a length of 3 hours.
- 1
-
Convert the
statechangesintointervals, i.e. from datetimes when a state changes to intervals where a state is consideredactive. - 2
-
Add a variable (
State.Brownby default) containing the Brown et al. (2022) phases. This function automatically extends the rows to include thepre-sleepphase.
Next, we combine the light dataset with the sleep data.
- 1
- Combine the two datasets
- 2
-
Get the start and end datetimes for each state from the
Intervalcolumn - 3
-
Assures that the times from the
sleep_datadataset are matched up with the light data, even though the light data uses theCentral European Time, whereas the sleep states use the defaultUTCtime zone. - 4
- Remove times that don’t relate to any state
# A tibble: 6 × 12
Datetime MEDI state State.Brown dawn
<dttm> <dbl> <chr> <chr> <dttm>
1 2025-09-24 14:00:31 6896. wake wake 2025-09-24 06:41:25
2 2025-09-24 14:01:31 6895. wake wake 2025-09-24 06:41:25
3 2025-09-24 14:02:31 6867. wake wake 2025-09-24 06:41:25
4 2025-09-24 14:03:31 6845. wake wake 2025-09-24 06:41:25
5 2025-09-24 14:04:31 6790. wake wake 2025-09-24 06:41:25
6 2025-09-24 14:05:31 6749. wake wake 2025-09-24 06:41:25
# ℹ 7 more variables: dusk <dttm>, photoperiod <drtn>, photoperiod.state <chr>,
# Reference <dbl>, Reference.check <lgl>, Reference.difference <dbl>,
# Reference.label <chr>7 Brown et al. (2022) recommendations
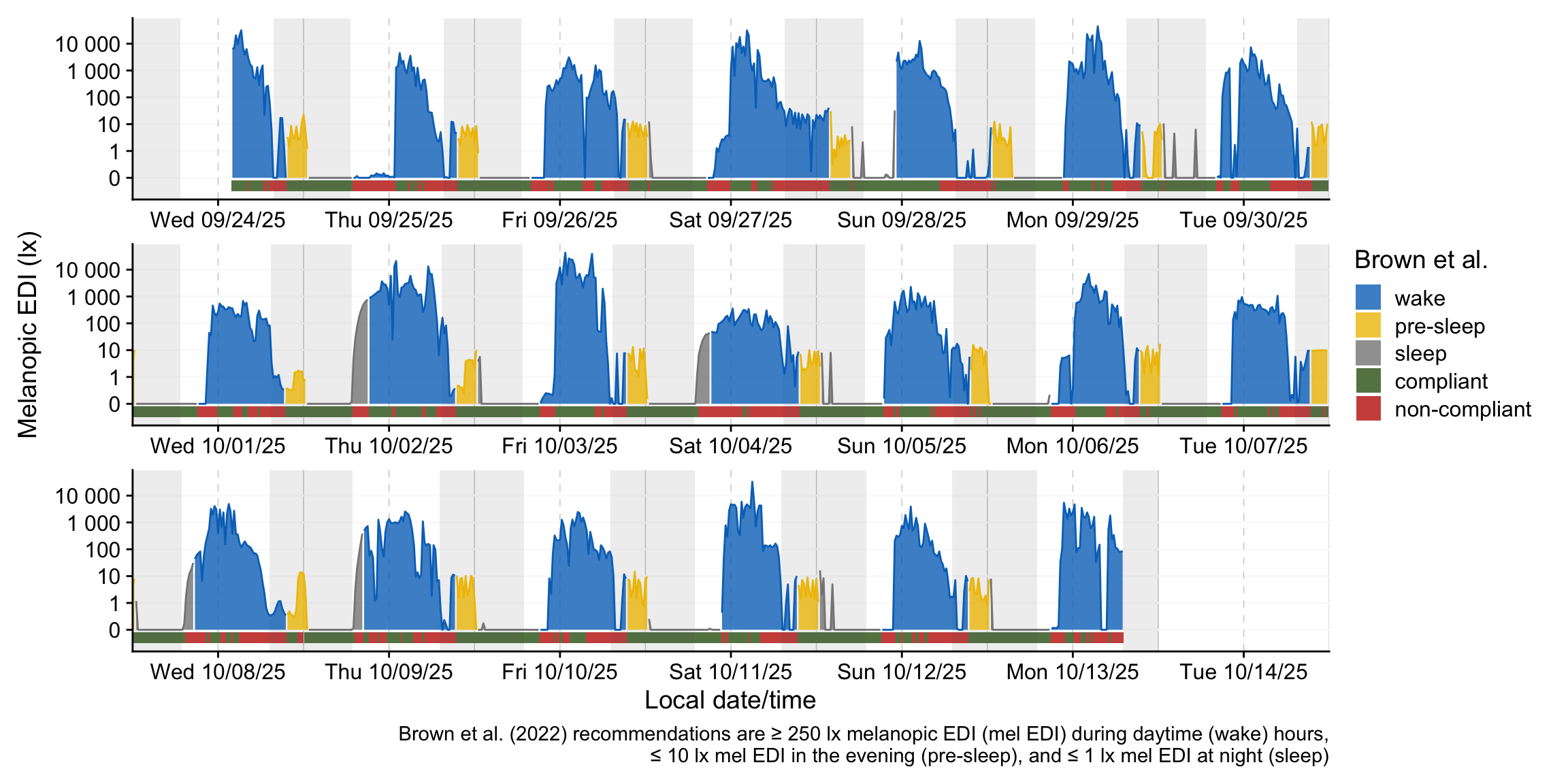
Let’s visualize the newly extended dataset with regards to the Brown et al. (2022) states.
color_palette <-
c(
wake = "#0073C2FF",
`pre-sleep` = "#EFC000FF",
sleep = "#868686FF",
compliant = "#658354",
`non-compliant` = "#CD534CFF"
)
data_ext |>
aggregate_Datetime("15 mins") |>
1 mutate(week = week(Datetime),
week = paste0("week: ", week),
Reference.check =
case_when(Reference.check ~ "compliant",
!Reference.check ~ "non-compliant"),
2 State.Brown = fct_expand(State.Brown, "compliant", "non-compliant") |>
fct_relevel("wake")
) |>
group_by(week) |>
3 gg_days(geom = "ribbon",
alpha = 0.8,
lwd = 0.5,
4 aes_col = State.Brown,
aes_fill = State.Brown,
group = consecutive_id(State.Brown),
5 x.axis.limits = \(x) {
Datetime_limits(x, length = ddays(6), midnight.rollover = TRUE)
}
) |>
gg_photoperiod(alpha = 0.1) |>
6 gg_states(Reference.check,
aes_fill = Reference.check,
7 ymax = -0.1, ymin = -0.5,
alpha = 1) +
labs(fill = "Brown et al.",
caption =
"Brown et al. (2022) recommendations are ≥ 250 lx melanopic EDI (mel EDI) during daytime (wake) hours,\n≤ 10 lx mel EDI in the evening (pre-sleep), and ≤ 1 lx mel EDI at night (sleep)") +
guides(color = "none") +
scale_fill_manual(values = color_palette) +
8 theme_sub_strip(background = element_blank(),
text.y = element_blank())- 1
- Grouping by week so that the plot is automatically sectioned into three rows
- 2
- The expansion of factor levels is purely so that the order in the legend is automatically kept in sensible order.
- 3
-
Setting the geom to
ribbonenables a filled band with the lower band set at 0. - 4
-
Coloring and filling by the Brown states will only look good, if each section is kept individually.
consecutive_id()ensures just this. - 5
- Manually setting the datetime limits ensures that each row is kept at 7 days
- 6
- We add a check whether the recommendations were fulfilled and color by it.
- 7
- Adjusting the height of the state bands ensures readability of the plot despite other color aspects.
- 8
- As the week grouping is just for row-setting, we are not interested in the strip and thus remove it.
We can further summarize the compliance to the recommendations in a table.
data_ext |>
group_by(State.Brown) |>
summarize(`Relative compliance` = sum(Reference.check)/n(),
`Relative duration` = n()) |>
mutate(`Relative duration` = `Relative duration` / sum(`Relative duration`),
State.Brown = fct_relevel(State.Brown, "wake")
) |>
arrange(State.Brown) |>
gt() |>
fmt_percent(decimals = 0)| State.Brown | Relative compliance | Relative duration |
|---|---|---|
| wake | 35% | 54% |
| pre-sleep | 89% | 12% |
| sleep | 95% | 33% |
8 Calculate metrics
In this section, we calculate some light metrics that we can correlate with sleep quality. We calculate these metrics not by the 24-hour day, but rather by wake-sleep rhythms.
8.1 Sleep-wake cycles
- 1
- Create a consecutive numbering per sleep-wake cycle. Beware if states are missing in other analyses!
- 2
- Format the sleep-wake cycles nicely
# A tibble: 6 × 15
# Groups: sleep.wake [1]
sleep.wake Datetime MEDI state State.Brown dawn
<fct> <dttm> <dbl> <chr> <chr> <dttm>
1 cycle 01 2025-09-24 14:00:31 6896. wake wake 2025-09-24 06:41:25
2 cycle 01 2025-09-24 14:01:31 6895. wake wake 2025-09-24 06:41:25
3 cycle 01 2025-09-24 14:02:31 6867. wake wake 2025-09-24 06:41:25
4 cycle 01 2025-09-24 14:03:31 6845. wake wake 2025-09-24 06:41:25
5 cycle 01 2025-09-24 14:04:31 6790. wake wake 2025-09-24 06:41:25
6 cycle 01 2025-09-24 14:05:31 6749. wake wake 2025-09-24 06:41:25
# ℹ 9 more variables: dusk <dttm>, photoperiod <drtn>, photoperiod.state <chr>,
# Reference <dbl>, Reference.check <lgl>, Reference.difference <dbl>,
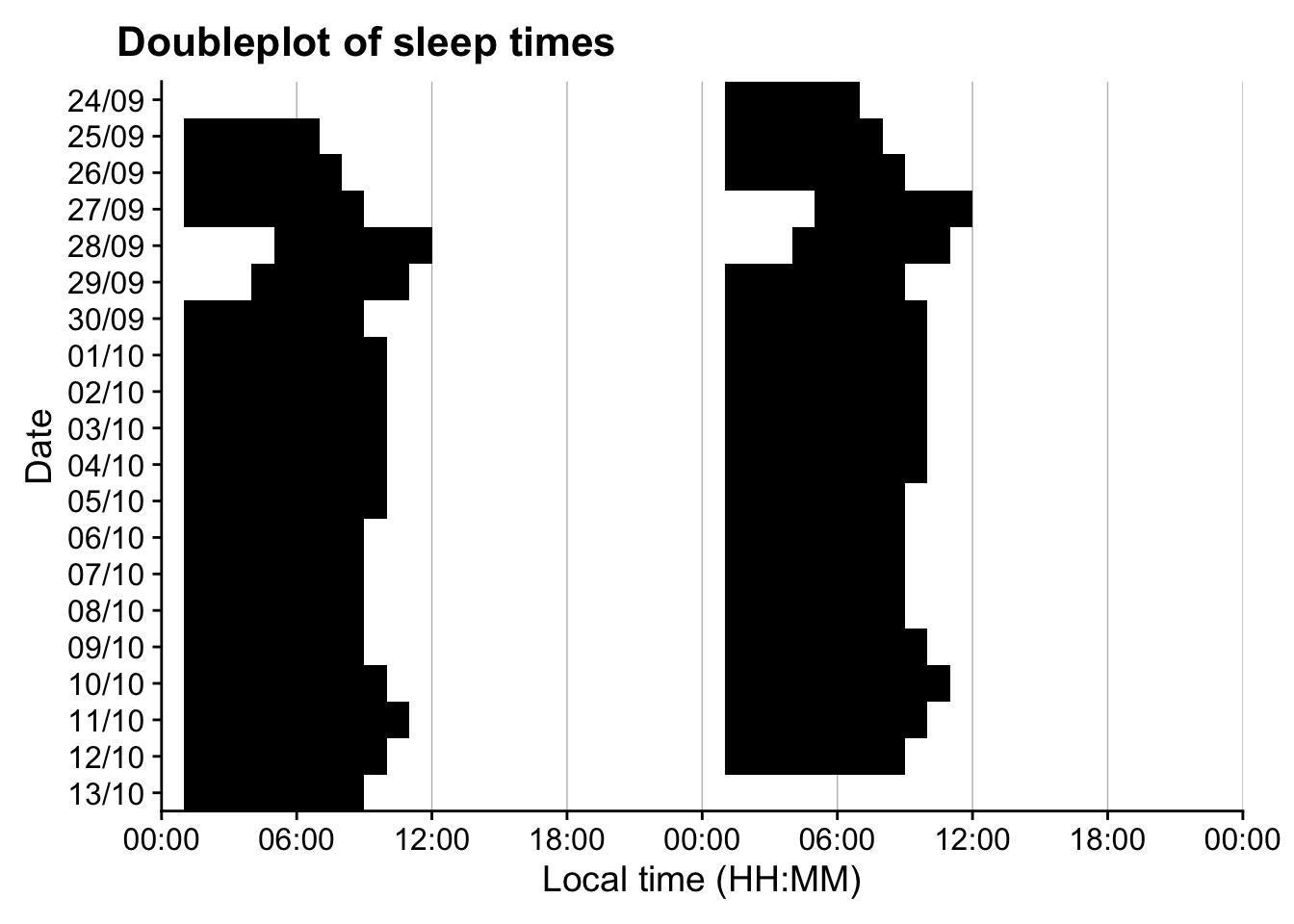
# Reference.label <chr>, Date <date>, state.count <int>Let’s look at the sleep times as a doubleplot.
data_sw |>
ungroup() |>
1 gg_heatmap(state == "sleep",
2 doubleplot = "next",
3 fill.remove = TRUE) +
scale_fill_manual(values = c("TRUE" = "black", "FALSE" = "#00000000")) +
guides(fill = "none") +
labs(title = " Doubleplot of sleep times") +
theme(
strip.background = element_blank(),
strip.text.y = element_blank(),
strip.placement = "inside")- 1
-
Create a logical as the main variable that is
TRUEwhen the state issleep - 2
- Showing the next day in the doubleplot
- 3
- Remove the default fill settings to provide your own
Again, we can look at the data as a table.
| sleep.wake | duration |
|---|---|
| cycle 01 | 60300s (~16.75 hours) |
| cycle 02 | 90000s (~1.04 days) |
| cycle 03 | 89100s (~1.03 days) |
| cycle 04 | 95400s (~1.1 days) |
| cycle 05 | 84600s (~23.5 hours) |
| cycle 06 | 77400s (~21.5 hours) |
| cycle 07 | 90000s (~1.04 days) |
| cycle 08 | 86400s (~1 days) |
| cycle 09 | 86400s (~1 days) |
| cycle 10 | 86400s (~1 days) |
| cycle 11 | 87300s (~1.01 days) |
| cycle 12 | 84600s (~23.5 hours) |
| cycle 13 | 86400s (~1 days) |
| cycle 14 | 85500s (~23.75 hours) |
| cycle 15 | 85500s (~23.75 hours) |
| cycle 16 | 89100s (~1.03 days) |
| cycle 17 | 91800s (~1.06 days) |
| cycle 18 | 81000s (~22.5 hours) |
| cycle 19 | 85500s (~23.75 hours) |
| cycle 20 | 36480s (~10.13 hours) |
Based on these results, we will remove cycle 01 and cycle 20, as they have only partial data.
8.2 Light exposure metrics
For the last step, we will calculate two relevant light exposure metrics, time above 250 lx mel EDI and dose. and add this information to the original states.
- 1
- This step relates the sleep cycles to the date where the wake-state starts.
- 2
- Adding the self-assessments to our summaries. The data (4) provides the right assignment.
We bring this information together into a comprehensive table. Variables that have no meaningful variance are removed.
summaries_sw |>
select(Date, sleep.wake, dose, duration_above_250,
wellbeing.morning, sunny.day, low.movement, performance,
fatigue, bedtime, sleepquality, sleepduration,
sleepinterruptions, sleepinterruptions.duration) |>
gt() |>
fmt_number(dose, decimals = 0) |>
fmt_duration(c(duration_above_250, sleepduration, sleepinterruptions.duration),
input_units = "seconds") |>
fmt_time(bedtime) |>
fmt(c(sunny.day, low.movement), fns = \(x) ifelse(x, "Yes", "No")) |>
tab_header("Overview of daily light exposure metrics and key questionnaire data") |>
tab_spanner("Light exposure", columns = c(dose, duration_above_250)) |>
tab_spanner("Self-report", columns = wellbeing.morning:fatigue) |>
tab_spanner("Self-reported sleep", columns = bedtime:sleepinterruptions.duration) |>
cols_label(sleep.wake = "Sleep-wake",
dose = "Dose mel EDI (lx·h)",
duration_above_250 = "Time above 250lx mel EDI",
wellbeing.morning = "Wellbeing next morning",
sunny.day = "Sunny day",
low.movement = "Low activity",
performance = "Performance",
fatigue = "Fatigue",
bedtime = "Bedtime",
sleepquality = "Sleep quality",
sleepduration = "Sleep duration",
sleepinterruptions = "Sleep interruptions",
sleepinterruptions.duration = "Interruption duration"
) |>
tab_footnote("Higher is worse", locations = cells_column_labels(c(8, 9, 11, 13, 14))) |>
sub_missing()| Overview of daily light exposure metrics and key questionnaire data | |||||||||||||
| Date | Sleep-wake |
Light exposure
|
Self-report
|
Self-reported sleep
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Dose mel EDI (lx·h) | Time above 250lx mel EDI | Wellbeing next morning | Sunny day | Low activity | Performance1 | Fatigue1 | Bedtime | Sleep quality1 | Sleep duration | Sleep interruptions1 | Interruption duration1 | ||
| 2025-09-25 | cycle 02 | 6,075 | 3h 8m | 4 | No | No | 4.0 | 2 | 00:30:00 | 2 | 6h 30m | 3 | 30m |
| 2025-09-26 | cycle 03 | 5,936 | 5h 16m | 4 | No | No | 3.0 | 1 | 00:15:00 | 2 | 7h | 3 | 30m |
| 2025-09-27 | cycle 04 | 32,863 | 5h 3m | 2 | No | No | 2.0 | 0 | 04:45:00 | 2 | 6h | 0 | 0s |
| 2025-09-28 | cycle 05 | 14,993 | 6h 28m | 2 | No | No | 2.0 | 3 | 03:30:00 | 4 | 7h | 2 | 5m |
| 2025-09-29 | cycle 06 | 42,373 | 5h 22m | 2 | No | Yes | 5.0 | 3 | 00:30:00 | 3 | 7h | 4 | 30m |
| 2025-09-30 | cycle 07 | 9,481 | 4h 46m | 4 | No | Yes | 3.0 | 1 | 00:15:00 | 2 | 6h | 5 | 2h |
| 2025-10-01 | cycle 08 | 2,832 | 4h 46m | 4 | No | No | 3.0 | 1 | 00:15:00 | 2 | 6h 30m | 4 | 1h 30m |
| 2025-10-02 | cycle 09 | 31,293 | 8h 3m | 3 | Yes | No | 3.0 | 0 | 00:15:00 | 4 | 4h 30m | 50 | 6h |
| 2025-10-03 | cycle 10 | 70,280 | 5h 29m | 3 | Yes | No | 4.0 | 2 | 00:15:00 | 4 | 4h | 80 | 6h 40m |
| 2025-10-04 | cycle 11 | 1,493 | 1h 33m | 4 | No | No | 4.0 | 2 | 00:30:00 | 3 | 6h 30m | 5 | 2h 30m |
| 2025-10-05 | cycle 12 | 4,222 | 4h 40m | 4 | No | — | 3.5 | 1 | 00:30:00 | 3 | 5h 45m | 5 | 2h |
| 2025-10-06 | cycle 13 | 9,246 | 4h 35m | 4 | No | Yes | 4.0 | 2 | 00:15:00 | 3 | 6h | 5 | 4h |
| 2025-10-07 | cycle 14 | 3,053 | 5h 13m | 4 | No | Yes | 3.0 | 1 | 00:15:00 | 3 | 6h 30m | 5 | 4h |
| 2025-10-08 | cycle 15 | 8,892 | 3h 25m | 4 | No | No | 3.0 | 1 | 00:15:00 | 3 | 4h 30m | 5 | 4h 30m |
| 2025-10-09 | cycle 16 | 6,585 | 4h 48m | 3 | No | No | 4.0 | 2 | 00:15:00 | 3 | 5h | 6 | 5h |
| 2025-10-10 | cycle 17 | 4,446 | 3h 18m | 4 | No | No | 3.0 | 1 | 00:15:00 | 3 | 4h | 10 | 6h |
| 2025-10-11 | cycle 18 | 24,397 | 4h 3m | 3 | No | No | 4.0 | 1 | 00:15:00 | 3 | 4h 30m | 10 | 5h 30m |
| 2025-10-12 | cycle 19 | 4,020 | 3h 14m | 3 | No | No | 3.0 | 1 | 00:15:00 | 3 | 4h 30m | 8 | 5h 30m |
| 1 Higher is worse | |||||||||||||
9 Correlations
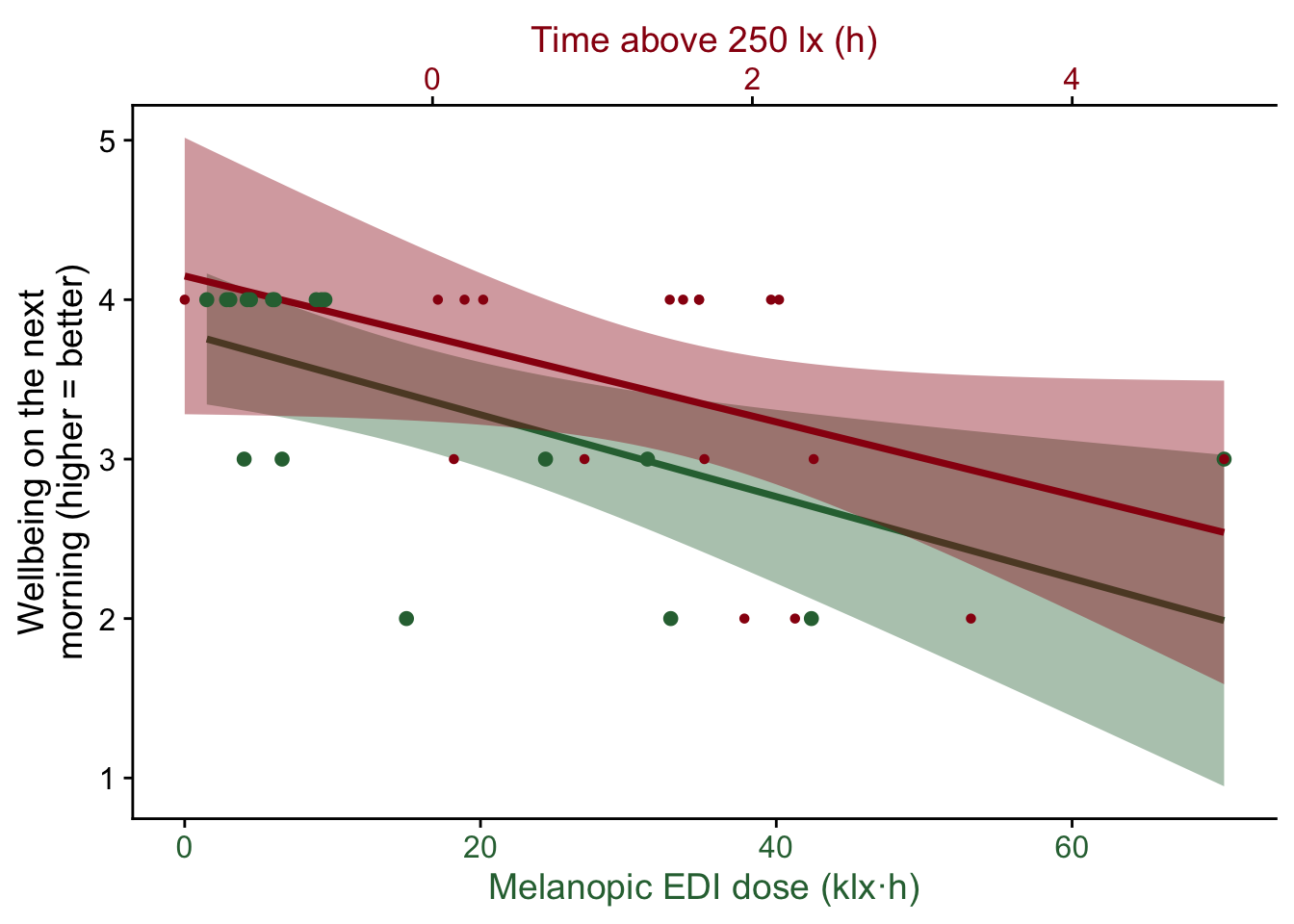
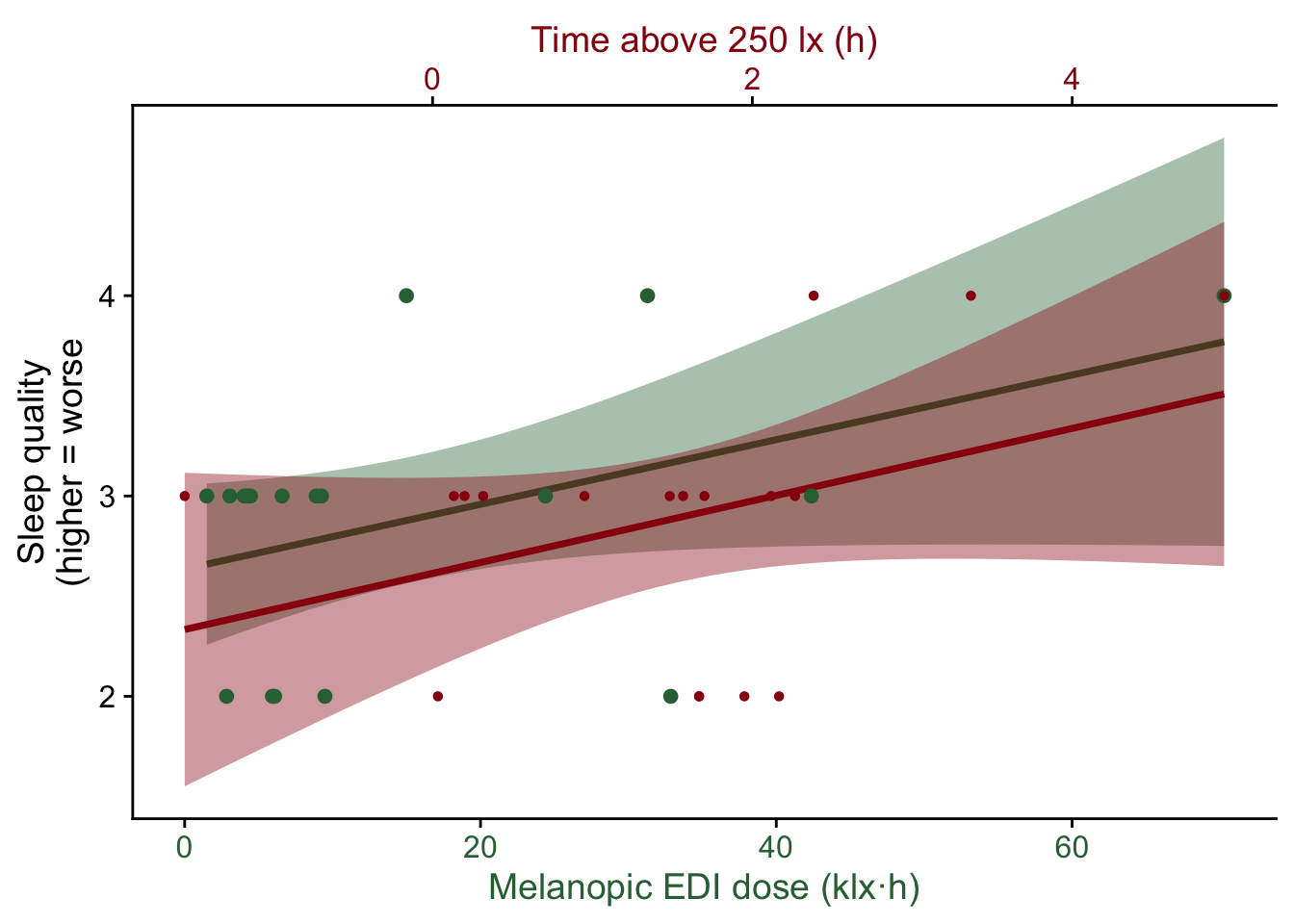
To visualize simple correlations between the self-assessments and the light exposure metrics we create a helper function:
time2hour <- function(x) x/3600
correlation_plot <- function(variable, label) {
summaries_sw |>
#prepare the light exposure variables and bedtime variable:
mutate(
bedtime = hms::as_hms(bedtime) |> as.numeric() |> time2hour(),
duration_above_250 = as.numeric(duration_above_250),
duration_above_250 =
(duration_above_250 - min(duration_above_250))/
(max(duration_above_250)-min(duration_above_250))*max(dose)/1000,
dose = dose/1000
) |>
#create the ggplot:
ggplot(aes(y = {{ variable }})) +
geom_smooth(method = "lm",
aes(x = dose),
col = "#2E6F40",
fill = "#2E6F40") +
geom_smooth(method = "lm",
aes(x = duration_above_250),
col = "#990011",
fill = "#990011") +
geom_point(aes(x = dose),
col = "#2E6F40") +
geom_point(aes(x = duration_above_250),
col = "#990011", size = 1) +
scale_x_continuous(
"Melanopic EDI dose (klx·h)",
sec.axis = sec_axis(~ (.*1000/70280*23400 - 5580)/3600,
name = "Time above 250 lx (h)")
) +
cowplot::theme_cowplot() +
theme_sub_axis_bottom(title = element_text(color = "#2E6F40"),
text = element_text(color = "#2E6F40")) +
theme_sub_axis_top(title = element_text(color = "#990011"),
text = element_text(color = "#990011")) +
labs(y = label)
}This allows us to easily look at the various parameters
Try these for yourself. Could there be a meaningful relationship?
correlation_plot(performance, "Performance during\nthe day (higher = worse)")
correlation_plot(performance.next,
"Performance on the\n next day (higher = worse)")
correlation_plot(sleepduration/3600, "Sleep duration (h)")
correlation_plot(fatigue, "Fatigue during the day\n(higher = worse)")
correlation_plot(fatigue.next, "Fatigue on the next day\n(higher = worse)")
correlation_plot(sleepinterruptions, "Sleep interruptions")
correlation_plot(sunny.day, "Self-assessed sunny day")
correlation_plot(low.movement, "Self-assessed low activity")10 Conclusion
Congratulations! You have finished this section of the advanced course. If you go back to the homepage, you can select one of the other use cases.